Welcome to the Mitchell Lab
At Brighton and Sussex Medical School, The University of Sussex
We’re tackling cancer with systems biology, in the beautiful city of Brighton, UK.
What we do
Our interdisciplinary approach aims to bring systems biology approaches to the clinic.
to represent cell signaling
to simulate disease
to test predictions
to improve treatments
Projects
Lab Alumni
Featured Publications
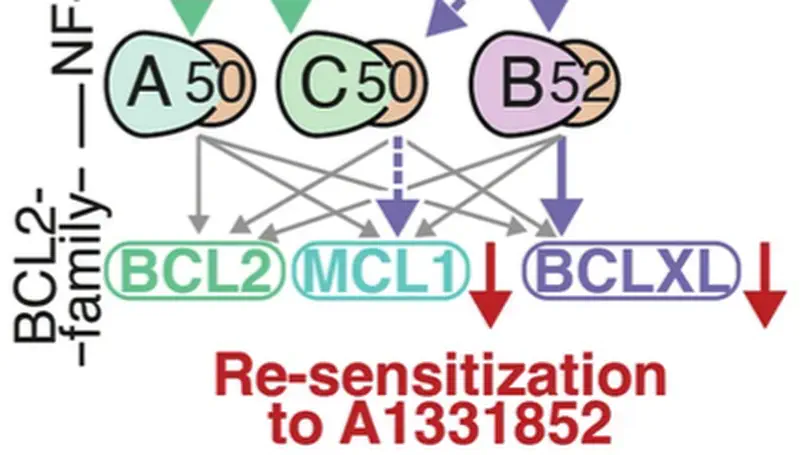
In Diffuse Large B-cell Lymphoma (DLBCL), elevated anti-apoptotic BCL2-family proteins (e.g., MCL1, BCL2, BCLXL) and NF-κB subunits (RelA, RelB, cRel) confer poor prognosis. Heterogeneous expression, regulatory complexity, and redundancy offsetting the inhibition of individual proteins, complicate the assignment of targeted therapy. We combined flow cytometry ‘fingerprinting’, immunofluorescence imaging, and computational modeling to identify therapeutic vulnerabilities in DLBCL. The combined workflow predicted selective responses to BCL2 inhibition (venetoclax) and non-canonical NF-κB inhibition (Amgen16). Within the U2932 cell line we identified distinct resistance mechanisms to BCL2 inhibition in cellular sub-populations recapitulating intratumoral heterogeneity. Co-cultures with CD40L-expressing stromal cells, mimicking the tumor microenvironment (TME), induced resistance to BCL2 and BCLXL targeting BH3-mimetics via cell-type specific upregulation of BCLXL or MCL1. Computational models, validated experimentally, showed that basal NF-κB activation determined whether CD40 activation drove BH3-mimetic resistance through upregulation of RelB and BCLXL, or cRel and MCL1. High basal NF-κB activity could be overcome by inhibiting BTK to resensitize cells to BH3-mimetics in CD40L co-culture. Importantly, non-canonical NF-κB inhibition overcame heterogeneous compensatory BCL2 upregulation, restoring sensitivity to both BCL2- and BCLXL-targeting BH3-mimetics. Combined molecular fingerprinting and computational modelling provides a strategy for the precision use of BH3-mimetics and NF-κB inhibitors in DLBCL.
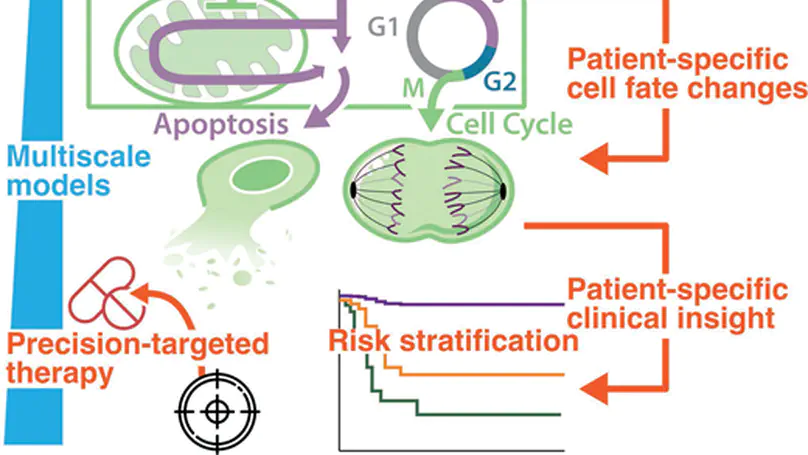
Decades of research into the molecular signalling determinants of B cell fates, and recent progress in characterising the genetic drivers of lymphoma, has led to a detailed understanding of B cell malignancies but also revealed daunting heterogeneity. While current therapies for diffuse large B-cell lymphoma are effective for some patients, they are largely agnostic to the biology of each individual’s disease, and approximately one third of patients experience relapsed/refractory disease. Consequently, the challenge is to understand how each patient’s mutational burden and tumour microenvironment combine to determine their response to treatment; overcoming this challenge will improve outcomes in lymphoma. This mini review highlights how data-driven modelling, statistical approaches and machine learning are being used to unravel the heterogeneity of lymphoma. We review how mechanistic computational models provide a framework to embed patient data within knowledge of signalling. Focusing on recurrently dysregulated signalling networks in lymphoma (including NF-κB, apoptosis and the cell cycle), we discuss the application of state-of-the-art mechanistic models to lymphoma. We review recent advances in which computational models have demonstrated the power to predict prognosis, identify promising combination therapies and develop digital twins that can recapitulate clinical trial results. With the future of treatment for lymphoma poised to transition from one-size-fits-all towards personalised therapies, computational models are well-placed to identify the right treatments to the right patients, improving outcomes for all lymphoma patients.
Recent Publications
Contact
- S.A.Mitchell@bsms.ac.uk
- Medical Research Building, University of Sussex, Falmer, East Sussex, BN1 9PX
- Enter the Medical Research Building (note this is behind the Medical Teaching Building) and call reception. We are on the second floor, up the stairs through the security door.
- Institution Website
- +44 (0)1273 678584
- ORCID iD
- Book a Meeting with Simon
- Google Scholar














